With Vaccination, Can We Resist the Evolutionary Force of Viruses?
COVID-19, caused by the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), has been a challenging pandemic in the 21st century1. The highly pathogenic SARS-CoV-2 has resulted in unprecedented and devastating socioeconomic impacts globally. At the very early stage of vaccine development, spike protein is regarded as a main antigenic target2. However, with the worldwide emergence of SARS-CoV-2 variants, particularly the one carrying spike protein variant, the effectiveness of COVID-19 vaccine against emergent variants becomes a major public concern.
A Key Determinant of Viral Infectivity and Transmissibility of SARS-CoV-2
COVID-19 has posed a tremendous threat to global public health since December 2019. Being an enveloped, positive-sense, single-stranded RNA virus, the viral envelope of SARS-CoV-2 is coated by spike, envelope, membrane proteins and the nucleocapsid proteins form the capsid to pack the genomic RNA (Figure 1)1. To mediate the viral entry, the spike proteins on the envelope of SARS-CoV-2 bind to angiotensin-converting enzyme 2 (ACE2) receptor on host cell membrane and is later endocytosed1-2.
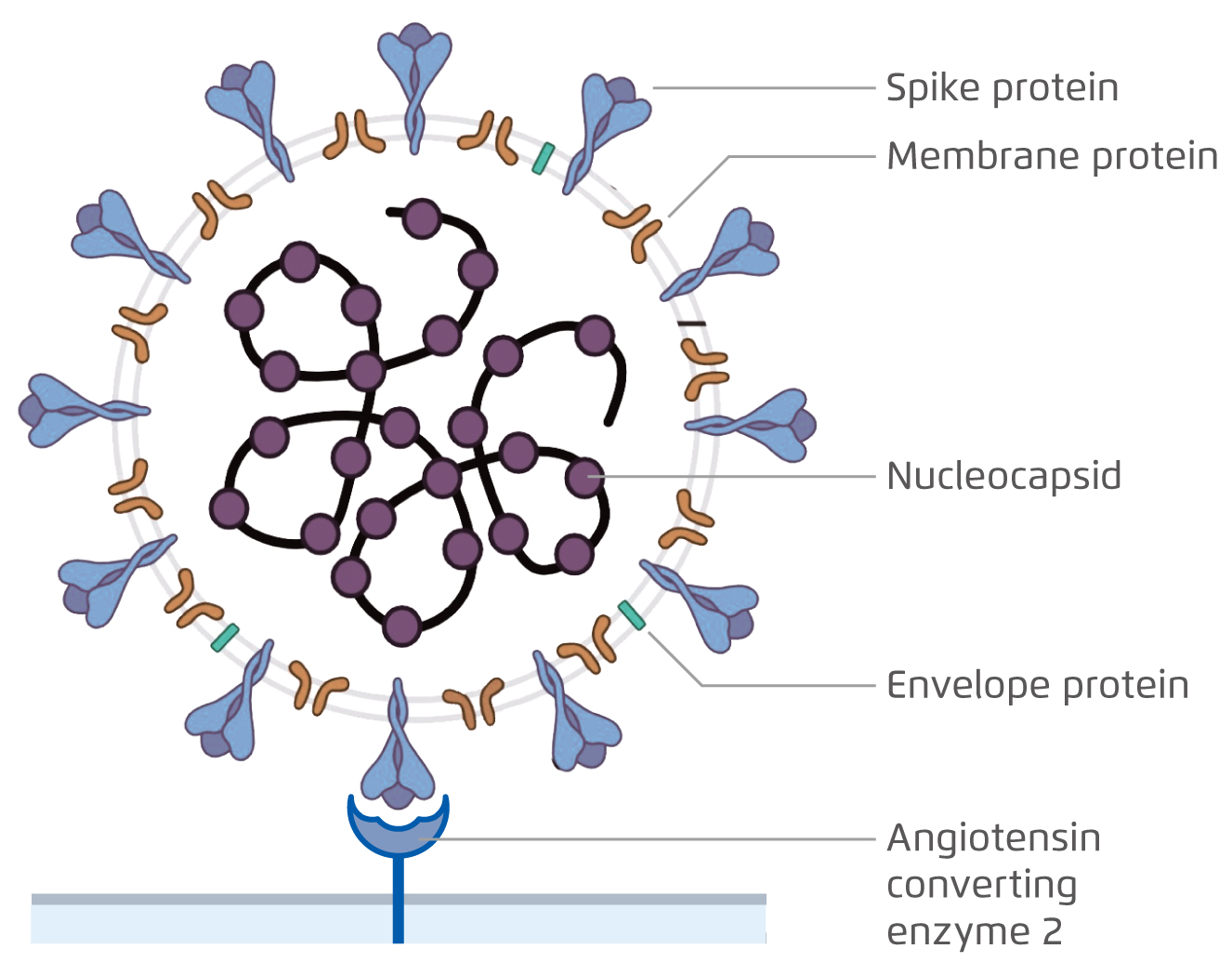
Figure 1. The virion structure of SARS-CoV-21.
Spike protein is a major determinant of viral infectivity and transmissibility in the host3. As antibodies that bind to the receptor-binding domain of the spike protein can prevent its attachment to the host cell, spike protein is later identified as a major therapeutic target for vaccine development against SARS-CoV-22. However, with the prior experience of vaccine resistance against influenza virus, considerable attention has focused on the antigenic evolution of the current circulating virus.
Underlying Mechanisms of Viral Antigenic Variation
It is not surprising to anticipate the occurrence of SARS-CoV-2 mutation as protein sequences in viral antigens constantly undergo mutation, which may result in antigenic drift or antigenic shift. Antigenic drift generally refers to the genetic mutation leading to the minor changes in antigens, whereas antigenic shift is the alteration in the antigenicity of a virus by reassortment between different viral strains and that results in an emergence of new strain of virus4,5. In fact, to ensure the effectiveness of vaccine against viral infection, investigating how drastically is the virus mutating is of unquestionable importance.
Influenza is a contagious respiratory illness that most likely occurs during the winter season and leads to an acute infection. Point mutation in the RNA segments coding for haemagglutinin or neuraminidase of influenza A virus can be occurred in antigenic drift and abolishes the binding of antibody5,6. Therefore, influenza virus may continuously escape from the recognition by the antibodies against the previous strains, which enables the novel strains to replicate and spread in a partially immune population after prior vaccination or infection5-6. Hence, annual review of the composition of seasonal influenza vaccine is required to prevent the infection caused by a new influenza strain7.
Compared with antigenic drift, antigenic shift raises a greater public health concern as genetic reassortment is involved and it may lead to a propagated pandemic by forming a new subtype of virus5. The outbreak of 2009 H1N1 Pandemic was a classic example of the antigenic shift with the genetic reassortments from the strains of human, avian and swine influenza viruses8. As a vast majority of people have little or no immunity against the novel virus, the emergence of new strain by reassortment has led to destructive consequences9. While influenza viruses keep on changing due to antigenic drift, antigenic shift occurs less frequently but its rarer occurrence is relatively worrying from a public health perspective owing to the lack of immunity against the new virus4.
Global Emergence of SARS-CoV-2 Variants
Different mutated variants of SARS-CoV-2 have alarmed the world about the viral mutation in SARS-CoV-2 genome, especially SARS-CoV-2 variant carrying spike protein variant G614 detected in this global pandemic10. As a result of the replacement of aspartic acid with glycine at position 614 of the spike protein of SARS-CoV-2, this variant has become the most prevalent form worldwide and global transition could be observed from the original D614 form to G614 variant (Figure 2)10-11.
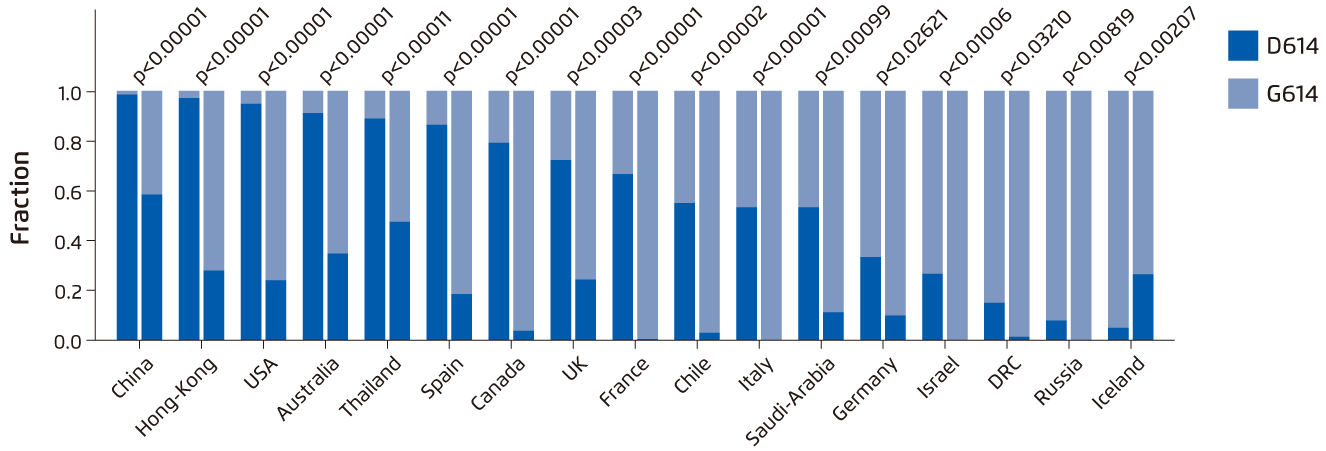
Figure 2. Paired bar charts which compare the fraction of sequences with D614 and with G614 for two time periods separated by a 2-week gap in March 202011.
Potential Outcomes of the Occurrence of G614 Variant
Preliminary study had reported that the increased infectivity and stability of the G614 virions were accompanied by an enhanced viral replication in human lung epithelial cells and primary human airway tissues12. Transmissibility of G614 variant may also be enhanced with the higher viral loads observed in the upper respiratory tract of COVID-19 patients and infected hamsters, whereas disease severity was not affected12,13.
Due to the urgency to develop vaccines, current COVID-19 vaccines in clinical trials are mainly based on the original D614 sequence and it raises a public concern on whether G614 variant may compromise the vaccine efficacy12. A serum neutralisation assay from the mice immunised twice at a 4-week interval with nucleoside-modified mRNA-LNPs encoding spike D614 sequence suggested that spike-based SARS-CoV-2 vaccines potentially neutralised the globally dominant G614 variant and vaccine efficacy were unlikely to be affected by G614 variant (Figure 3)14. These findings may alleviate the concerns regarding the current efforts to develop an effective SARS-CoV-2 vaccine and underscores the importance of evaluating the current vaccines against G614 variant.
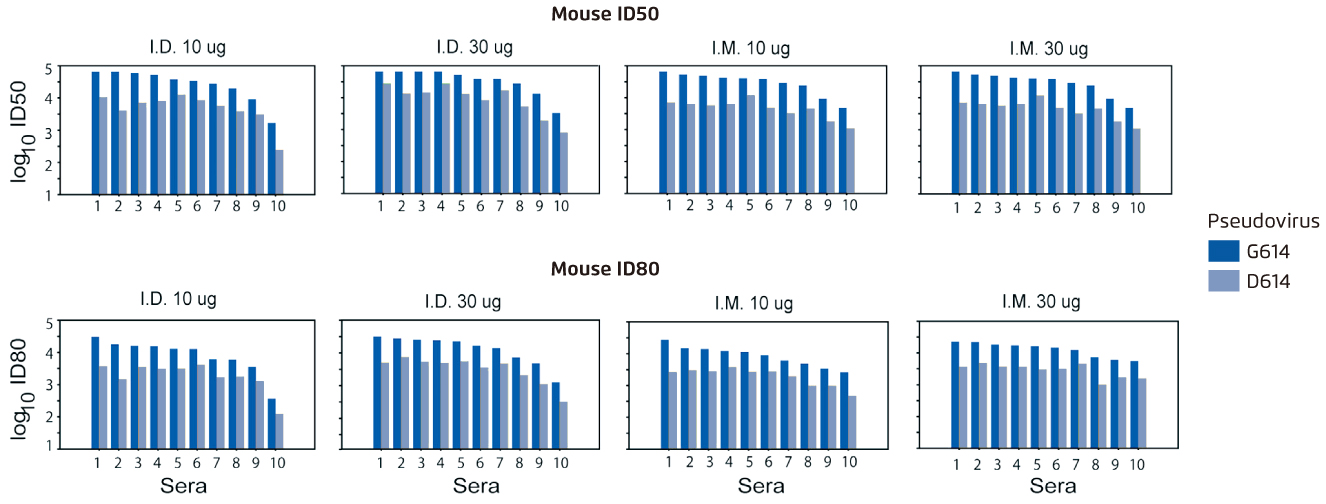
Figure 3.
A serum neutralisation assay which the sera (10 mice/group) obtained 4 weeks after the second immunisation were tested for neutralisation against pseudoviruses with the D614 and G614 variants of spike14. Higher log10 values of ID50 and ID80 indicate increased neutralisation activity. I.D.=intradermal; ID50=inhibitory dose to reduce infection by 50%; ID80=inhibitory dose to reduce infection by 80%; I.M.=intramuscular
Importance of SARS-CoV-2 Strain Surveillance
Although multiple vaccines against SARS-CoV-2 have been administered, different SARS-CoV-2 variants have been discovered in South Africa and the United Kingdom in the meanwhile15. Further investigations are urgently warranted to understand the potential impacts of different SARS-CoV-2 variants on their viral properties and their responses toward the vaccines developed15. Also, close monitoring of circulating viral strains and accumulating mutations within SARS-CoV-2 genome is of incontestable importance in vaccine development.
References
1. Li YD, et al. J Biomed Sci 2020;27:104. 2. Krammer F. Nature 2020;586:516-527. 3. Li Q, et al. Cell 2020;182:1284-1294.e9. 4. Centers for Disease Control and Prevention. How the Flu Virus Can Change: “Drift” and “Shift”. Available at https://www.cdc.gov/flu/about/viruses/change.htm. (Accessed on 18 January 2021). 5. Naeem A, et al. J Med Virol 2020;10.1002/jmv.25759. 6. Treanor J. N Engl J Med 2004;350:218-220. 7. Centre for Disease Control and Prevention. Selecting Viruses for the Seasonal Influenza Vaccine. Available at https://www.cdc.gov/flu/prevent/vaccine-selection.htm. (Accessed on 18 January 2021). 8. Cheng VC, et al. Clin Microbiol Rev 2012;25:223-263. 9. Centers for Disease Control and Prevention. 2009 H1N1 Pandemic (H1N1pdm09 virus). Available at https://www.cdc.gov/flu/pandemic-resources/2009-h1n1-pandemic.html. (Accessed on 18 January 2021). 10. Volz E, et al. Cell 2021;184:64-75.e11. 11. Korber B, et al. Cell 2020;182:812-827.e19. 12. Plante JA, et al. Nature 2020;10.1038/s41586-020-2895-3. 13. Baric RS. N Engl J Med 2020;383:2684-2686. 14. Weissman D, et al. Cell Host Microbe 2021;29:23-31.e4. 15. World Health Organisation. SARS-CoV-2 Variants. Available at https://www.who.int/csr/don/31-december-2020-sars-cov2-variants/en/. (Accessed on 18 January 2021).